Research
The TIGRR Lab is a new research center focused on marsupial conservation and restoration. The research focuses on three main research themes:
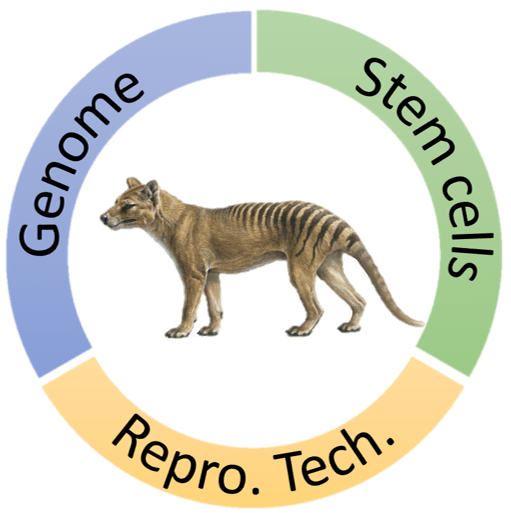
- Marsupial genomics and improvement of the thylacine genome.
We aim to produce the best quality genome possible to learn more about the biology of this unique marsupial and to aid in its de-extinction. This involves the sequencing of many different specimens to determine species variation. We will also compare the thylacine genome to that of the dunnart (a closely related dasyurid marsupial) to determine the quantity and scope of edits required to create a thylacine-like marsupial cell. - Refinement of methods for the derivation and maintenance of marsupial stem cells.
We will establish methods that could be used to biobank diversity in our Australian marsupials. This will also help protect against species loss from habitat destruction and other stressors such as predation or competition with invasive pests. Marsupial stem cells would also be used to pioneer gene editing for conservation efforts in extant marsupials and provide a template cell for the de-extinction of the thylacine. - Development of assisted reproductive techniques (ART) in marsupials.
Techniques such as IVF and cloning are very well established in placental mammals, but completely lacking in marsupial. These technologies could be used to revive lost diversity in extant marsupial populations, greatly aid in marsupial breeding programs and will be required to one day facilitate the transformation of a thylacine-like cell into a living organism.
Marsupial Conservation
Australia holds the world record for the most mammal extinctions. The latest report from IUCN’s Red List shows we are the 5th worst country for animal extinctions globally, and the 6th worst for the number of species listed as endangered and critically endangered.
Generating marsupial stem cells provides a means for biobanking (freezing and banking stem cells) to safeguard against the loss of species and of genetic diversity. With recent bushfires, this is an essential conservation tool to develop for marsupials. The Kangaroo Island dunnart (pictured below) is just one of many species that suffered a dangerous population decline due to the recent fires.

Gene-editing marsupial genomes could save endangered species. The northern quoll is classified as endangered. This is largely because they prey on introduced cane toads, which are toxic and deadly to animals that eat them. Animals that coevolved alongside cane toads in South America have naturally evolved resistance to the toxin. With our gene editing and marsupial assisted reproductive technologies in hand, we can engineer cane toad toxin-resistant quolls. This has the potential to save the northern quoll from extinction, with the added benefit that they might help control the cane toad population that is impacting many other native mammals, reptiles and amphibians.


These shorter-term conservation projects involve the development of technologies that will ultimately be essential for the longer-term project of thylacine de-extinction, providing a mutual interdependence that sustains research momentum.
Thylacine de-extinction
Step 1 – complete
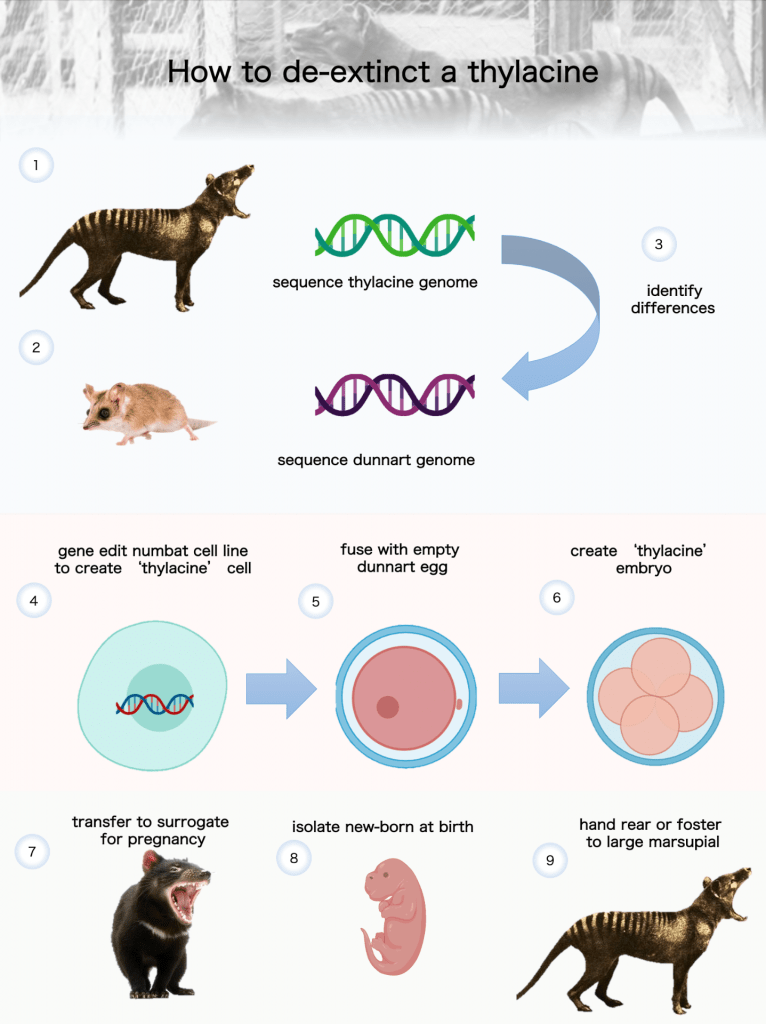
The genome provides the complete blueprint or set of instructions on how to build a thylacine. It is an essential first step in any de-extinction project, and the likelihood of de-extincting the animal is completely reliant on the quality and accuracy of that genome.
To date, the thylacine genome is the highest quality extinct genome for any species.
However, this resource can still be improved. Advances in genome sequencing technology since the first thylacine genome in 2017 provides new options to further enhance the quality and build of the genome.
Step 2 – complete
We now have a platinum level genome for one of the thylacine’s closest living and most easily bred relatives, the fat-tailed dunnart (Sminthopsis crassicaudata). This species will provide the living cells and genomic template that will be used to create a functional Tasmanian tiger genome, and later a complete specimen.
Step 3 – in development
We are seeking additional funding to see how widely our methods for making marsupial stem cells can be applied. We would like to establish methods that work in all marsupials. With this tool in hand, we can effectively bio-bank diversity in ‘at-risk’ marsupial populations and protect against the loss of species that are threatened or endangered.
This is a large bioinformatics (computational) project to compare both genomes and identify all the differences that would potentially need to be edited into the numbat genome to create a ‘thylacine’ cell.
Step 4 – in development
We currently have some funding from the Australian Research Council (ARC) to develop methods to derive marsupial stem cells. We have achieved this for our model marsupial species, the fat-tailed dunnart, in which a lot of the techniques needed for our thylacine de-extinction will be developed.
Steps 5-7 – in development
This requires the development of assisted reproductive technologies (ART) for marsupials. These techniques are required in order to use living stem cells to make an embryo and then successfully transfer it into a host mother’s uterus. Procedures like enucleating dunnart oocytes to accept material from a “thylacine” cell, embryo transfer and culture will all be advanced markedly going forward to benefit de-extinction and conservation.
Steps 8-9 – the future
De-extinction efforts for marsupials have a distinct advantage over other mammals. All marsupials give birth to tiny young regardless of the size of the adult, so the fetus outgrowing the mother’s uterus is not a concern. Marsupials then complete development in the pouch while sucking milk. This can be done with bottle feeding or in a host pouch – marsupials are very tolerant of fostering pouch young of other species.
Thylacine developmental biology
We are exploring the thylacine genome to determine which pieces are the most critical for its unique development. This work involves large scale genome wide analyses and comparisons with other marsupials (its closest living relatives) and with the canids (dogs) with whom it shares a striking resemblance. This is a phenomenon known as convergent evolution.
We have shown that thylacine and wolf represent the best known example of convergent evolution ever described in mammals.
This striking similarity in body shape – and especially the shape of the skull – enables us to ask the simple question: if two very distantly related species evolve to look the same, is that reflected by similar changes in their genome?
The short answer to this compelling question in developmental biology – is yes! And the regions of the genome that we have identified though these analyses are informing us about the biology of the thylacine, its unique development and the most important regions we would need to edit for a de-extinction project.
This research is uncovering fundamental answers about how mammals evolve and which regions of the genome drive key developmental events.

Thylacine population structure and genetic health
If a future thylacine population is to thrive in the wild it will be necessary to understand the genetic health of the population prior to extinction to ensure a robust breeding program into the future. Our previous work has shown limited genetic diversity within the Tasmanian thylacine population prior to extinction. However, our future work will sequence the genome of multiple individuals to try to recapture some genetic diversity within any future population.

Development of the dunnart as a lab-based marsupial model species
Despite their unique biology, marsupial resources are lagging behind those available for placentals mammals. The fat-tailed dunnart (Sminthopsis crassicaudata) is a laboratory based marsupial model, with simple and robust husbandry requirements and a short reproductive cycle making it amenable to experimental manipulations.
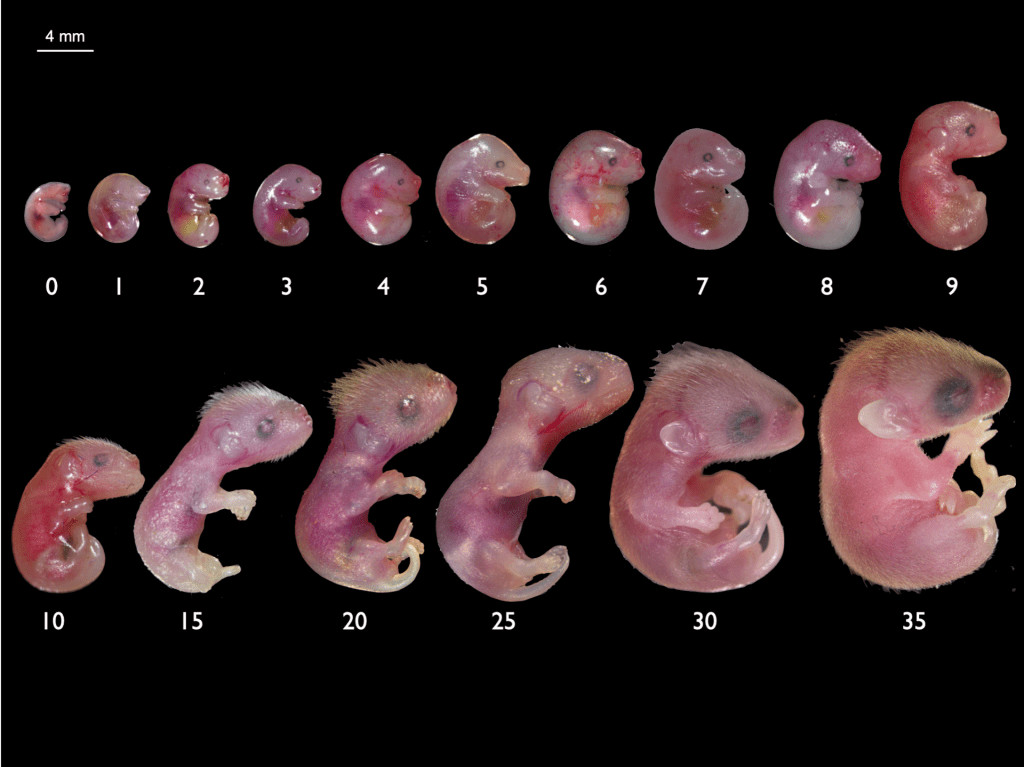
Gross morphology of the fat-tailed dunnart throughout postnatal days of development in
the pouch. https://www.nature.com/articles/s42003-021-02506-2
We have developed a detailed staging series for the fat-tailed dunnart, focusing on their accelerated development of the forelimbs and jaws. This study provides a fundamental resource for future studies exploring genome manipulations in marsupials.
Want to get involved in our research?
More information
Related Publications
Peel E, Frankenberg S, Hogg CJ, Pask A, Belov K. Annotation of immune genes in the extinct thylacine (Thylacinus cynocephalus). Immunogenetics 2021; 73:263-275.
Newton AH, Weisbecker V, Pask AJ, Hipsley CA. Ontogenetic origins of cranial convergence between the extinct marsupial thylacine and placental gray wolf. Commun Biol 2021; 4:51.
Menzies BR, Renfree MB, Heider T, Mayer F, Hildebrandt TB, Pask AJ. Limited genetic diversity preceded extinction of the Tasmanian Tiger. PLoS ONE 2012; 7(4) e35433.
Cook LE, Newton AH, Hipsley CA, Pask AJ. Postnatal development in a marsupial model, the fat-tailed dunnart (Sminthopsis crassicaudata; Dasyuromorphia: Dasyuridae). Commun Biol 2021; 4:1028.
Newton AH, Pask AJ. CHD9 upregulates RUNX2 and has a potential role in skeletal evolution. BMC Mol Cell Biol 2020; 21:27.
Newton AH, Pask AJ. Evolution and expansion of the RUNX2 QA repeat corresponds with the emergence of vertebrate complexity. Commun Biol 2020; 3:771.
Feigin CY, Newton AH, Pask AJ. Widespread cis-regulatory convergence between the extinct Tasmanian tiger and gray wolf. Genome Res 2019; 29:1648-1658.
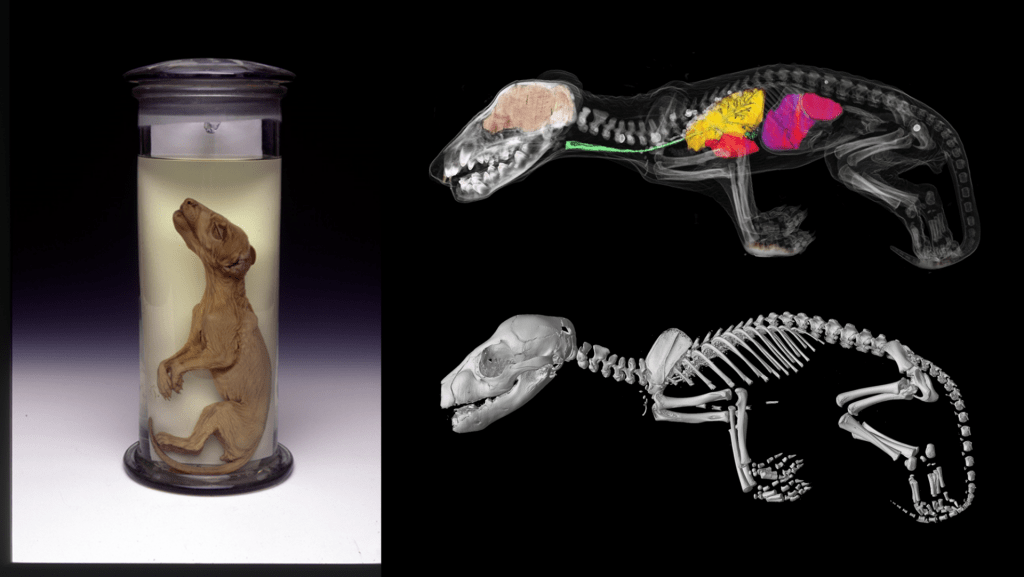
Newton AH, Spoutil F, Prochazka J, Black JR, Medlock K, Paddle RN, Knitlova M, Hipsley CA, Pask AJ. Letting the ‘cat’ out of the bag: pouch young development of the extinct Tasmanian tiger revealed by X-ray computed tomography. R Soc Open Sci 2018; 5:171914.
Feigin CY, Newton AH, Doronina L, Schmitz J, Hipsley CA, Mitchell KJ, Gower G, Llamas B, Soubrier J, Heider TN, Menzies BR, Cooper A, et al. Genome of the Tasmanian tiger provides insights into the evolution and demography of an extinct marsupial carnivore. Nat Ecol Evol 2018; 2:182-192.